Outline of MM-GBSA scoring project for protein design (12-26)
[These notes are also copied on the academic wiki since the other collaborative projects are listed there. We have included it here due to its connection to the MM-GBSA sirtuin project.]
RC: Karthik, the NSF sirtuin proposal was oriented towards protein design, but there is no protein design in the projects we have planned. See below for such a project. This can take the place of the project originally proposed using the python protein design code, which we considered a little too complicated for the students. If you agree on offering this project, please start making the slides based on the points I have made below (a couple of slides on background, a couple on approach). The Boas and Harbury paper mentioned below should be in the dropbox - Brad was using it.
MM-GBSA rescoring of the sequence libraries generated in PNAS papers. No resampling of sequences to start - use those already generated in PNAS. RC will provide the files.
Background and purpose: While glidescore performs well at rank-ordering sequences for a given ligand, it does not provide accurate binding free energies. Evidence in literature (e.g. Boas in protein design, others EK has found in drug design) suggests MM-GBSA can also rank order sequences or ligand poses. (MM-GBSA is most useful in drug design for rank ordering cogeneric series of ligands for the same receptor – a problem where ensemble averages for the unbound ligand are important – but in protein design the ligand remains fixed and the receptor changes). There is also evidence that MM-GBSA can predict binding free energies with appropriate experimental calibration, unlike Glidescore. (Here, experimental calibration would require a catalytically inactive substrate analog, and we will not do experiments at this point. “Cogeneric series” used for calibration is a series of sequences. Experiments could later be done on sirtuins.) Accurate calculation of sequence entropies and evolutionary temperatures (assuming Km \approx Kd) -- see the slide on sequence entropies in above slide set -- requires accurate prediction of relative binding free energies. Hence we will rescore the sequences obtained in the PNAS papers using MM-GBSA, with the aim of ultimately being able to computationally predict the changes in binding free energy of the substrate due to mutations, analogously to the manner in which MM-GBSA is being used by EK to predict binding affinities of cogeneric inhibitors.
Approach: Our prior work suggests MM-GBSA may not be as effective as glidescore in rank ordering sequences. Students would read papers on use of MM-GBSA for pose sampling (EK has noted that there are papers in the literature that use MM-GBSA for pose sampling - please try to find them), as well as protein design (Boas) -- these papers suggest that MM-GBSA --is-- effective at rank-ordering, and write a script that would do energy minimization and scoring (using both MM-GBSA and Glidescore) of sequence lists provided by us for several enzymes including beta-lactamase. For MM-GBSA, a single ensemble calculation is needed for each ligand. If MM-GBSA does not rank order the sequences like glidescore, students analyze the breakdown of contributions to glidescore and MM-GBSA scores, scale vdW radii and other free parameters, and explain why the rank orderings differ.
Outcomes: Could result in paper jointly between PMC-AT and CMU, to be followed with a paper applying the python protein design code for sequence sampling. Any code written for ensemble MM-GBSA calculations could be integrated with the python protein design code.
Could connect KM's work for thesis to protein design.
Would give us more time to get cloud resources and associated codes for sampling up and running.
Slides for both Anna talk could be largely borrowed from previous talks, with KM making a few more for the proposal above, while refining his GBSA slides.
Esp useful if MD slides not fully developed, or if issues with MD accuracy.
RC Comments on 9-10 sirtuin draft
I have posted here my comments and plan for our future revisions (in which I will also be engaged) for both computation and experiment. Let us start with the * * points, then the * points, and then EK and I will work to address other computational revisions/additions while XG will revise experimental sections.
Question for discussion (please comment): Due to length should we consider splitting paper into 2? Do not want this to compromise quality of any one paper or to delay submission. Would allow us to include more of our results on computation. Biochemistry is not a computation-heavy journal. Possible breakup:
1) fluorescence assay / docking (no binding affinity estimation or iso-NAM). Possible journal: Biochemistry.
2) continuous assay/MM-GBSA binding affinity calculations, and iso-NAM expts and computation. Possible journal: JMB.
We will not do any more work either experimentally or computationally if we choose to break up. We can decide while making the corrections below, most of which are needed either way.
Computational part
These are to be addressed jointly by RC/EK. Priorities for EK are marked with **. Others will be worked on joinrly
1) * *pg 29-34: Clarify specification of which induced fit protocol was used to get these binding affinities. 4/20, 8/10 of docked structures – refer to the same docking protocol?
2) Pg 34: Justification of customized induced fit protocol – better sampling; if energy are lower the sampling is justified. But mention that enhanced sampling could be improved by an iterative algorithm
3) Pg 42: Conclusion – add proposal to prescreen potential activators by MM-GBSA docking followed by experimental screening
4) * *Do we have the MM-GBSA binding affinities for iso-NAM?
5) Any comments on analysis of uncompetitive inhibitor mechanisms using similar techniques?
6) * *Bottom of pg 10: further description in methods section on why docking scoring functions rather than MM-GBSA are used for sampling poses (be aware of length).
7) * *Related to 6), a breakdown of the components of a Glidescore and comparison to the MM-GBSA scoring function.
8) (RC) pg 42: Brief discussion of stationarity issues w MCMM and possible future improvements
9) (RC/EK; later) (pg 42) Description of equations for ensemble calculations to be put in computational methods section
10) pg 29-35: Ensemble calculation results: showing <E> estimate is improved by ensemble sampling but the T\Delta S correction is small/negligible
11) pg 14: Some brief description of the plop backbone sampling method desirable (this is not an exhaustive search)
12) pg 11-15: Order of paragraphs in methods section – alternates between induced fit/customized induced fit, etc
13) pg 10; Some more discussion in methods section on why ensemble calculations on bound states as not as important as / are more challenging than ensemble calculations on the free ligand (methods for sampling joint protein/ligand degrees of freedom, differences in density of states)
14) (RC) pg 23-29: introduce calculation of Kd from Km, kcat experimental results and compare to experimental predictions
15) * *Discuss normalization of MM-GBSA binding affinities. EK to post a paper on this. We will probably just mention that normalization can’t be done here but could be done with a larger dataset on sirtuins/sirtuin inhibitors.
Let us 1st address the * * parts then RC/EK will jointly address the remaining issues and write up together (RC will have some input on how to write up the remaining pts).
Experimental part
1) (pg. 39-40) Any reason to include further details on iso-NAM activation based on “Mechanism of SIRT1 activation” document? There is already some discussion here. Add discussion on base exchange reaction rates only if splitting into two papers
2) Clearer indication of when the fluorescence vs continuous assays are being used – which plots correspond to which assay?
3) Pg 39-40: abrupt transition between inhibition theory and iso-NAM discussion. Also, are we contrasting iso-NAM with allosteric activators like resveratrol here? Should be stated.
(RC): On the latest manuscript draft: a) we should briefly mention how plop optimizes side chains, since the difference with respect to minimization may not be clear to readers. Make sure to refer to rotamer optimization and explain what a rotamer is. b) It is stated that the induced fit protocol only optimizes side chains (not backbones). Is this right? In any case, we used loop optimization in our modified protocol. Do we refer to the loop prediction algorithm in plop anywhere in the paper? c) Excluded volume constraints are mentioned. I don't believe they were used for SIRT3. If they were used for Sir2, we could perhaps be more clear as to precisely where.
Raj
August 23, 2012 (Thu)
(RC) Enumerate the roles of the computational (sampling) tools described (in the wiki 8-10 update and in the latest [[#|draft]]) in future [[#|studies]] aimed at answering questions posed in the tasks in “Sirtuin Kinetics Paper [[#|draft]] 5-16-2012” that we decided not to do in this paper.
(EK) Here are the hypotheses from the “Sirtuin Kinetics Paper [[#|draft]] 5-16-2012”. Most of them have been answered, at least with docking and MM-GBSA binding affinity estimates. The ones that could not be done involve hypothesis 3 because the current methods with docking do not allow testing of induced structural changes beyond the localized environment of the docking area (through Induced Fit or other loop / side chain minimization).
(RC) To what extent have these been integrated into the draft?
- Hypothesis 1a: (SIRT3) Simulation to test if NAD+ preferentially docks to AC pocket.
- until very recently, we were going to scrap this because of the difficulty with docking NAD+ into the AC pocket. I have finally been able to dock NAD+ into the AC pocket as well as the AB pocket described on the Aug. 21 update. I also calculated MM-GBSA estimates for binding affinity, and NAD+ does preferentially dock to the AC pocket with lower calculated binding affinity. So we can include this in the current paper.
- Hypothesis 1b: (Sir2) Simulation to test if NAD+ preferentially docks to AC and AB pocket equally well.
- Done. MM-GBSA estimates indicate approximately equal binding affinity between the AB and AC pockets. Thus, hypothesis supported.
- Hypothesis 2 and 3: (treated together because similar)
- Hypothesis 2: (SIRT3) Simulation to test if NAD+ does not bind or binds with a much lower score to SIRT3 when NAM present in pocket C. This would be competitive inhibitor situation in which the NAM is blocking NAD+ binding to the C pocket, and AB binding of NAD+ is not as favored as NAD+ binding to the AC pocket. If this hypothesis is correct, NAM binding to the C pocket does not induce a structural change that sterically blocks NAD+ binding to the AB pocket, but changes the configuration of, for example, hydrogen bonds, to make NAD+ binding less favorable.
- Hypothesis 3: (SIRT3) a parallel to hypothesis 2, except than in this case that NAM binding to the C pocket induces structural changes that make NAD+ binding to the AB pocket sterically hindered. Hypothesis 3 is a quasi allosteric inhibition by NAM.
- We can include hypothesis 2 in the current paper. Results to support this hypothesis 2: MM-GBSA simulations indicate that NAD+ binds with a higher affinity to AC vs. AB. Nicotinamide docks to the C pocket in SIRT3. Thus, when nicotinamide is blocking the C pocket, there is competitive inhibition, as AB pocket binding is higher in energy. Although we don't know what structural changes could be induced by the inhibitor, we can still use the current results in support of the proposed mechanism.
- To test structural changes we do an MD trajectory with the nicotinamide pre-docked into the C pocket. We relax the system with MD. After the system has stabilized, we compare the AB binding pocket before and after. MM-GBSA could be used. Additionally, we could use FEP or umbrella sampling method to calculate the binding affinity of NAD+ in the relaxed system with the nicotinaimde bound to the C pocket.
- I suspect that there will be no large structural changes upon binding of the nicotinamide inhibitor. It is not a large molecule like the peptide substrate or the NAD+, and probably does not induce large structural changes. Thus, I hypothesis that nicotinamide simply blocks AC binding without inducing any structural changes
- Note: I do need to do additional docking simulations to test nicotinamide binding to the C pocket in the apo-enzyme (without NAD+, but with and without the acetyl-lysine peptide bound). This are not the above mentioned MD simulations, but more simulations to test if nicotinamide binds to the C pocket when NAD+ or the acetyl-lsyine peptide substrate are not there. Recall that the peptide binding or the NAD+ binding induces significant structural changes, which change the A, B, and C pockets.
- Hypothesis 4: (Sir2) NAD+ binds with equal affinity to AB and AC pockets.
- NAM is a noncompetitive inhibitor of NAD+. Unlike in SIRT3, the non-competitive inhibition can be explained with NAD+ docking/binding with approximately equal binding affinity to the AB or AC binding pocket. Thus, if NAM occupies pocket C, NAD+ will equally bind to pocket AB, but no reaction will proceed.
- The MM-GBSA binding affinity predictions supported this hypothesis.
- Hypothesis 6: (SIRT3 and Sir2) iso-NAM binds to the C pocket preferentially.
- Docking for SIRT3 and Sir2 both confirm this hypothesis
- Hypothesis 7: (SIRT3 and Sir2) iso-NAM binds to the B pocket preferentially.
- No, iso-NAM was shown to bind to the C pocket preferentially.
(RC) Mention the possibility of induced fit docking of the Sirtris uncompetitive inhibitor of SIRT3; mention that since this is an uncompetitive inhibitor, based on the definition of uncompetitive from XG's section, there must be a conformational change that occurs upon NAD+ binding to the AC pocket.
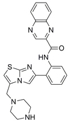
(EK) Raj, by this, you mean SRT1720, shown here? It is a quinolinecarboxamide compound that acts as a potent, reversible and NAD+ uncompetitive and AceCS2 (Acetyl-CoA synthetase 2) peptide-competitive allosteric inhibitor of SIRT3 activity. In uncompetitive inhibition, the inhibitor (SRT1720) binds exclusively to the ES (protein-NAD+) complex, rather than to the free enzyme form (apo-protein). When the substrate (NAD+) binds, a conformational change occurs in the enzyme which forms or unmasks the inhibition site. The resulting enzyme-substrate-inhibitor complex is catalytically inactive. So the answer to your statement is: Yes, there must be a conformational change that occurs upon NAD+ binding to the AC pocket. It may be possible to search for SRT1720 docking sites with a docking program or with induced fit, using the SIRT3-NAD+ complex. However, since there is no crystal structure of this complex, it is more challenging to create the proper structure using various computational techniques. So far I have docked NAD+ into SIRT3 by minimizing certain sterically clashing side chains and backbone atoms starting from the trapped thio-imidate intermediate. This is the closest structure to the SIRT3-NAD+ complex. But this intermediate may not contain the proper binding site for the SRT1720 inhibitor, because that very inhibitor probably prevents this intermediate imidate from forming.
(RC): Yes the reason I mentioned this is that we are discussing tools (e.g. induced fit) that allow us to determine conformational changes that occur upon NAD+ binding. These same tools are undoubtedly important in studying the mechanism of action of uncompetitive inhibitors, due to the definition above of uncompetitive inhibitors. We now know a little bit about the conformational changes that occur upon NAD+ binding. We should mention this in the paper (while indicating the caveat that larger scale conformational changes may occur that are responsible for the mechanism of action of SRT1720).
Aug. 21, 2012
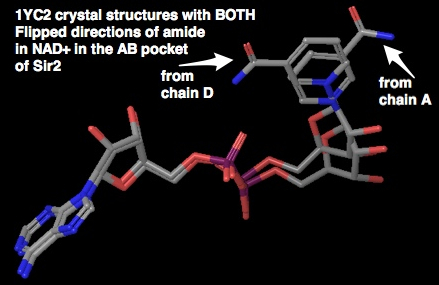
A note on the AB pocket conformation of NAD+: it can adopt two flipped conformations. Previous descriptions of the NAD+ in the AB pocket crystal structure showed the amide only flipped towards the phosphates. There is another chain in the multimeric crystal structure of 1YC2 which has the amide of the nicotinamide end of NAD+ flipped the other way. This is important because some of the docked structures have the amide flipped out, which we now know is also seen in the crystal structures.
Docking NAD+ into AB and AC pockets after PRIME refinement: no constraints
Maestro Project Name: redock.after.prime
- PRIME refinement of sterically clashing side chains with the NAD+ ligand in the AB conformation.
- Did standard protein preparation wizard on 3GLT (SIRT3 with ADPR bound to the AceCS2 peptide containing a thioacetyl lysine)
- Deleted the NAD+ intermediate and the bound peptide substrate, which are co-crystallized in 3GLT
- Superimposed backbone of 1YC2 chain A (Sir2Af2) onto 3GLT
- Extracted NAD+ from the 1YC2:A superposition, and incorporated it into the 3GLT protein structure. This is NAD+ in the AB pocket conformation
- Ran PRIME loop refinement only for the residues (and +/- 1 to 2 neighboring residues) that had steric clashes with the superimposed NAD+. These refined residues were:
- A:157 to A:160 AND A:320 to A:324
- a subsequent additional loop side chain minimization was done because docking failed with just the above side chain / loop refinements. Side chain atoms of VAL366 were overlapping with an atom on the adenine of the NAD+ (this is on the opposite end of the NAD+ from the nicotinamide and is firmly in the A pocket) This may have happened because of protein movement to accommodate the trapped ADPR intermediate, which has a slightly different conformation than NAD+ in the AC pocket before reaction with the acetyl lysine. Thus, A:365 to A:367 (VAL366 and its neighbors) were minimized with PRIME.
- The PRIME options:
- default serial loop sampling method
- refined side chains within 6.0 Å
- all other options default; no constraints
- Docking Grid Setup: No Constraints; No excluded volumes; default Grid parameters; Flexible ligand
- Grid enclosed by 17 Å box on a side centered on the NAD+ superimposed from 1YC2:A
- Scaling factor 1.0 (none) with partial charge cutoff of 0.25
- Glide Docking
- SP mode; standard docking
- Completely flexible ligand. starting ligand conformation is NAD+ from AB conformation from 1YC2:A
- Add epik state penalties to score; use input partial charges
- partial charges calculated by epik for NAD+ from the AB conformation from 1YC2:A in H2O solvent with pH 7.0 +/- 2.0
- scaling factor of 0.8 for non-polar parts of ligand with partial charge cutoff of 0.15
- output at most 512 poses per ligand
- all other settings are default for Glide
- Simulation name: glide-dock_SP_24_noConstraints_flexible_pv1
- Results:
- 8 out of top top 10 (based on the emodel glide score) docked the NAD+ into the AC pocket (see figure below)
- 4 out of the top 20 (based on the emodel glide score) docked the NAD+ into the AB pocket (see figure below)
- An important result here is that no constraints or excluded volumes were used. However, the NAD+ in the AB conformation from 1YC2:A (Sir2Af2) was used as a template to refine the side chains and backbone of the above described residues which sterically clashed with the AB conformation, which could be considered a type of constraint. However, observing both the AC and AB conformations from this simulation without further constraints on the protein or the ligand is a result that we have been searching for. The best Emodel score for the top ranked conformation (in the AC pocket) is -174.3, while the best emodel score for NAD+ in the AB pocket is -154.9. Emodel numbers are not as reliable as MM-GBSA, shown below.
- MM-GBSA results:
- AC pocket:
- The best AC pocket result is -107.9
- for top 20, results are -73 to -107.9 for those conformations in approximately the AC pockets, with a median around -89 (better than the AB docking)
- AB pocket
- MM-GBSA calculated binding affinities for the top 4 poses out of the 20 highest ranked (from emodel scores) closest to the AB conformation of NAD+ from 1YC2:A (RMSD shown)
- RMSD MM-GBSA
1.82 -84.4
2.17 -71.5
2.18 -75.9
2.48 -60.8
This preliminary result agrees with our hypothesis that NAD+ would bind with higher affinity to the AC pocket than the AB pocket for SIRT3, unlike in Sir2, where MM-GBSA calculations indicated that NAD+ probably binds with approximately equal affinity. - RC (8-21): Do we have the Prime backbone/sidechain reorganization energies handy? Let us add those to the MM-GBSA binding energies since there is an energetic cost to getting the protein reorganized to accept these poses. For the paper, I would prefer to refer to plop algorithms rather than Prime, since it is academic code.
- AC pocket:
- TODO
- run with Glide XP. XP may improve the ligand positioning in the receptor, which may affect the MM-GBSA results.
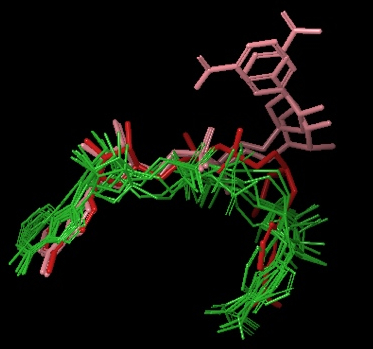
Above Figure: 8 out of top 10 (based on emodel glide score) docked the NAD+ into the AC pocket. Green are these 8 molecules. Red is the NAD+ in the AC pocket from the co-crystallized structure of 1YC2:B. Pink are the 2 structures from 1YC2 (chains A and D) with NAD+ in the AB pocket. The amide from the nicotinamide is pointing in both directions.
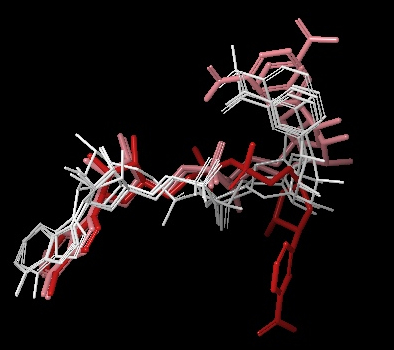
Above Figure: 4 out of the top 20 (based on the emodel glide score) docked the NAD+ into the AB pocket. White are these 4 molecules. Similar to the other figure with AC pocket docking from this same simulation, Red is the NAD+ in the AC pocket from the co-crystallized structure of 1YC2:B. Pink are the 2 structures from 1YC2 (chains A and D) with NAD+ in the AB pocket. The rank order of these 4 structures was 11, 13, 17, and 18 with RMSD to the superimposed 1YC2:A NAD+ of 2.18, 1.82, 2.17, 2.48 Å, respectively.
Docking of iso-nicotinamide and nicotinamide into SIRT3
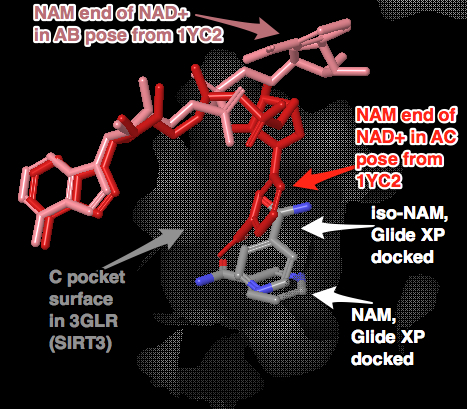
Above Figure: Docking of iso-NAM and NAM without constraints into 3GLR, human SIRT3 with acetyl-lysine AceCS2 peptide, but no NAD+ or intermediate co-crystallized. The same PRIME refined structure as used in the above NAD+ docking was used. Glide XP produced only the above shown poses, which were not exactly aligned with the nicotinamide end of the backbone aligned 1YC2:B NAD+. Note that the iso-NAM has the amide end pointing in a different direction than the NAM. It is not clear why the two inhibitors adopt different conformations from the superimposed nicotinamide end of the NAD+. It could be that the side chains (or backbone?) in the C pocket in the crystal structures from SIRT3 are slightly different than that of the C pocket in Sir2 which has NAD+ co-crystallized in the AC pockets. Recall that when NAM was docked into this Sir2 structure, the NAM almost exactly aligned with the nicotinamide end of the NAD+. At a minimum, the iso-NAM and NAM docked in the general C pocket volume, which is an open void allowing much wiggle room for these small molecules. I could do a more careful comparison between the C pockets of Sir2 with NAD+ in the AC pockets and this SIRT3.
A similar docking simulation was done with 3GLT (that has the ADPR thio-intermediate co-crystallized), which resulted in slightly poorer results with respect to RMSD to the reference nicotinamide of the reference NAD+ from 1YC2:B. The top ranked Glide XP iso-NAM and NAM still docked within the region of the C-pocket, but farther away from the reference. The pocket was slightly deformed because of the trapped thio-acetyl ADPR intermediate. For this reason, I chose to use 3GLR, which only has the acetyl-lysine peptide co-crystallized. Unlike for Sir2, no crystal structures with NAD+ bound (not intermediate) are available, so 3GLR was the next best option.
Comment on priorities among remaining tasks listed in the above document (esp iso-NAM docking). XG's draft has a section on iso-NAM. Indicate whether you did iso-NAM docking. Does it bind in the C pocket? See my comments on XG's Mon tasks regarding iso-NAM.
Yes, iso-NAM docking into SIRT3 was done. See Docking Simulations wiki page from Aug. 21. Yes, it binds in the C pocket when using Glide XP.
You asked us to expend our current conclusions to indicate rational design of more effective activators than iso-NAM based on computational studies - e.g., prediction of reaction rates for iso-NAM and NAM dissociation from the C pocket. The possible key to isoNAM's activation of Sir2/SIRT3 in the presence of the more potent nicotinamide inhibitor is isoNAM's ability to antagonize nicotinamide binding to the sirtuin. Nicotinamide inhibition has been shown to be linked not only to binding in the C pocket, but also to reversal of the reaction of the intermediate imidate to the re-formation of the NAD+. IsoNAM is an unreactive isostere, that potentially binds to the C pocket, but cannot participate in the reversal of the reaction because of the shifted location of the nitrogen on the pyridine ring of nicotinamide. This shift disrupts covalent binding to the imidate intermediate, preventing reformation of NAD+.
One path to computationally design these types of inhibitors is to balance increased C-pocket affinity with design of compounds that are non-reactive with the imidate intermediate. A more potent C-pocket binding, but non-reactive species to the imidate, must also be able to leave upon the next reaction cycle - either by movement in the C-pocket due to the deaceylated peptide leaving or the the product 2’-O-acetyl-ADPR leaving. Alternatively, one could imagine a high affinity activator leaving when the next NAD+ that bound to the AB pocket shifts it's nicotinamide end, booting out the activator. While these possible mechanisms are conjecture, this activator must, at some point dissociate from the C pocket so the NAD+ can react with the acetyl-lysine peptide.
You asked us to contrast iso-NAM and [[#|resveratrol]] as Sirtuin activators. It is not clear to me how resveratrol works, so this is difficult to answer.
Aug 16, 2012
Identify flexible loop regions that could be sampled instead of minimized in AB pocket of SIRT3. Sample with PRIME.
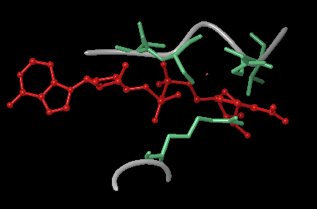
From SIRT3, 3GLT, the above figure shows the important flexible loop regions around the B pocket that are responsible for the steric clashing. The red molecule is the NAD+ in the AB conformation (not co-crystallized, but structurally aligned), while the aqua are the main side chains interfering with the B pocket. The grey backbone trace shows important loop regions that can be sampled. The top loop is GLY319 to GLH325, and the bottom is PHE157 to PRO160. The method involved superimposing the AB structure NAD+ (red molecule) onto the 3GLT crystal structure, then using PRIME to sample around the the NAD+. PRIME sampled the loops and moved the backbone and side chains out of the B pocket. The next step would be to re-dock NAD+ into this PRIME optimized structure. A similar method was used for Sir2.
Outline the steps in the Induced Fit Docking (IFD)
The below outline of IFD is from: Sherman, W., Day, T., Jacobson, M.P., Friesner, R.A., and Farid, R. (2006). Novel Procedure for Modeling Ligand/Receptor Induced Fit Effects. Journal of Medicinal Chemistry 49, 534–553.
The overall procedure (outlined in Figure 1) has four steps:
- initial softened-potential docking into a rigid receptor to generate an ensemble of poses;
- challenge in 1st step is to generate at least one reasonably docked pose for the ligand (independent of the score it receives). Without a plausible initial guess for the ligand pose, any attempt to predict reorganization of the protein structure is unlikely to succeed in the context of a limited allotment of CPU time.
- sampling of the protein for each ligand pose generated in the first step;
- challenge in the 2nd step is predicting the low energy receptor conformation for a correct ligand pose, starting from the “plausible” initial guess generated in the first step
- reducing of the ligand into low energy induced-fit structures from the previous step;
- challenge in the 3rd step is to generate low energy ligand conformations when presented with the correct receptor conformation.
- scoring by accounting for the docking energy (GlideScore), and receptor strain and salvation terms (Prime energy).
- challenge in 4th scoring step lies in properly ranking the complexes such that the top ranked pose correctly predicts the ligand/ receptor structure.
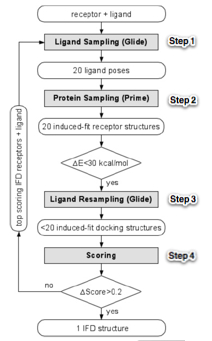
Figure 1: IFD flowchart. ∆E is the energy gap from the lowest energy structure
Comment on the challenges inherent in identifying activators via docking methods (e.g., one may need to know the rate of dissociation of iso-NAM,NAM)
A part of the docking studies was to explain the activation by iso-NAM in the presence of an inhibitor, and to explain the differences between Sir2 and SIRT3 activation by iso-NAM. One of our hypothesis about how iso-NAM activates Sir2 in the presence of the NAM inhibitor is that iso-NAM and NAM both bind to the C pocket, competitively, but iso-NAM has a higher dissociation rate (it leaves easier). Some of the challenges in using docking to identify activators:
- one may want to know the relative dissociation rates of the inhibitor and activator. This constant is related to the binding affinity:
- k1 (rate of dissociation) AB ---> A+B
- k2 (rate of association) A+B --->AB
- k2 / k1 = [A][B] / [AB] = Kd where Kd is the dissociation constant
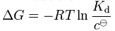 where ∆G is the binding affinity
where ∆G is the binding affinity- docking may not correctly rank order the potential activators by dissociation rates. Docking with MM-GBSA has a difficult enough time rank ordering for binding affinity.
- The true binding affinities and dissociation rates are not necessarily the simple binary AB--> A + B situation described above. The system has at least 3 molecules: NAD+ cofactor, the inhibitor, NAM (or the activator iso-NAM), and the protein. This complicates the picture.
- Searching for high affinity ligands to the C pocket may not be the best strategy. Iso-NAM may be a weaker inhibitor of Sir2 than NAM, which is why it becomes an activator in the presence of NAM. Thus, the highest ranked or highest predicted binding affinity ligands may end up being strong inhibitors, rather than activators.
Aug. 9, 2012
Discussion of past problems with SIRT3 AB docking and future direction.
Docking of NAD+ into the AB pocket of SIRT3 is challenging. A number of techniques with traditional docking (Glide) and Induced Fit, including various constrains, have been tried without success. Details of these simulations have been provided previously. This document focuses on causes of post problems and future directions with SIRT3 and the AB pocket. There are twomain approaches in moving forward: (1) additional traditional docking and induced fit, and, (2) advanced molecular dynamics techniques. For these techniques, two significant issues are sufficient ligand pose sampling and sidechain/backbone sampling to alleviate steric clashing.
An alternative argument is that AB docking in SIRT3 it too high in energy or sterically hindered in the real system. This result is partially supported by the experimental data that nicotinamide is a competitive inhibitor of NAD+. The non-competitive mechanism of Sir2 is that NAD+ can dock into the AB pocket while the inhibitor occupies the C pocket. When this inhibitor leaves, the nicotinamide of NAD+ rotates from the B pocket into the C pocket for productive binding. For SIRT3, competitive inhibition implies that NAD+ docking to the AB pocket is unfavorable or impossible when the inhibitor is present in the C pocket. Thus, the simulations failing to dock into the AB pocket are expected. Barring severe steric hinderance making docking impossible, we would like to dock the NAD+ into the AB pocket, even if it is a high energy pose, so that we can get the MM-GBSA estimated binding affinity in the AB pocket to compare to the AC pocket binding.
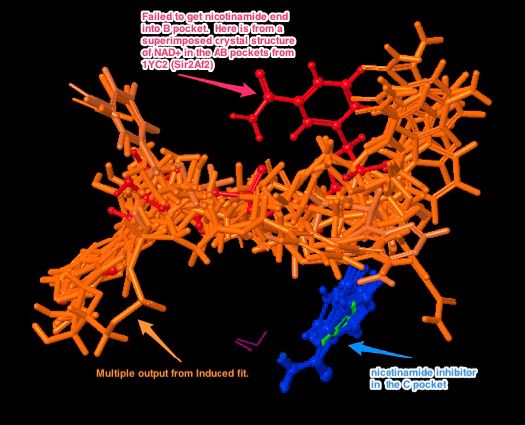
Before I discuss possible solutions for SIRT3 simulations, here is a summary of the difficulties in previous simulations. The below left picture, previously described, shows residues in the B pocket in SIRT3 (PDB:3GLT; aqua residues) sterically hindering the nicotinamide end of NAD+ (molecule in the center colored by element), compared to residues in the B pocket in Sir2 (Sir2Af2, PDB:1YC2; red residues). This picture shows that at least 3 residues are sterically blocking the NAD+ from docking into the B pocket. A critical issue is whether these clashes can be alleviated via side chain sampling and ligand pose sampling, or does it require larger backbone movement which Induced Fit will not accommodate. There may be backbone movements necessary that are larger than what can be done in Induced Fit. The below right picture shows the possible backbone movements that may be necessary to open up the B pocket. I created this by running a minimization with NAD+ harmonically constrained in the AB pockets (pose from the co-crystallized structure of Sir2 with NAD+ in the AB pocket), and minimized the structure of the protein around the NAD+. The backbone needed to move > 1.7 Å. This test was not comprehensive, because, for example, minimization does not properly sample side chain rearrangements. However, it suggests that backbone movement is a real problem.
Interestingly, when I ran Induced fit again, using this new structure which had an active site minimized to accommodate the harmonically restrained NAD+ in the AB pose, the protocol still failed to re-dock NAD+ into the AB pocket. This result suggests there is a ligand pose sampling problem, whereas previously there may have been both a ligand pose sampling and a backbone/side chain sampling problem. What I suspect is that the scoring function rejects poses in the B pocket due to unfavorable energy. In this case, we know that the NAD+ should dock in the AB pockets, because that is the starting structure that was minimized. However, the scoring function focuses on poses with better scores. I tried to increase the number of output poses to > 1000 (after clustering), thinking that the AB pocket pose would show up, but just at a worse GlideScore. It did not. It is possible to increase the number of output poses even more to 10,000 or 100,000, and rank order these poses based on RMSD to the input AB pocket pose (Induced Fit and Glide can do this rank ordering). Note that this ranking does not affect the type of poses that are output based on the same internal sampling and GlideScore thresholds. It simply sorts the output.
There are probably backbone/sidechain sampling issues, as well, for this energy minimized structure. If Induced Fit comprehensively sampled the backbone and side chains, it should find a receptor structure into which the NAD+ can dock into the AB pocket with a reasonable GlideScore that would appear in at least the 1st 1000 output structures. But because of limited backbone/sidechain sampling, this receptor structure and ligand pose is not found.
Induced Fit's limited constraint control is frustrating. Glide has the following constraints that are not available in Induced Fit: Atom/residue positional constraint, hydrophobic constraints (hydrophobic regions of receptor that are important), excluded volumes, and ligand torsional constraints. We could alter the Induced Fit script to include other Glide constraints like excluded volumes, atom position constraints, ligand torsion constraints. Excluded volumes and atom positional constraints would focus the sidechain/backbone and pose sampling to the AB pocket. Induced Fit is a Python script that iteratively and alternately calls Glide then PRIME. In theory, the rest of the Glide constraints should work, but have just not been coded into the Induced Fit script. Note that adding these constraints may be more complex, as it may complicate the grid generation during each iteration. On the downside, it may take some time for me to hack the code. Schrodinger may be interested in improving their program.
Raj asked how excluded volumes focus pose sampling. It's not that the use of excluded volumes generates more cumulative poses. Rather, it allows the poses that are generated to focus on away from excluded volumes to volumes of interest. Without the excluded volumes, poses in those interest areas may not pass the threshold GlideScore, and would automatically be pruned from the tree before additional pose sampling.
Simulated annealing molecular dynamics is an alternative approach. We've been focusing on using docking programs to estimate binding affinity. Docking is typically done when the ligand pose and/or (for induced fit) active site side chain arrangement is unknown. However, our case involves knowing approximately the pose (in the AB pocket), but not knowing the side chain and backbone rearrangement. A more appropriate simulation would be to run molecular dynamics with simulated annealing (or something similar) to allow the active site to relax around the AB pose. Some flexibility would be allowed in the ligand. After the trajectory has relaxed and stabilized, a collection of low energy conformations could be input into MM-GBSA.
The next [[#|step]] would be to use a more sophisticated molecular dynamics protocol. With MM-GBSA, a single structure of the ligand and receptor is used to estimate binding affinity. The more sophisticated MD protocols use an ensemble of structures from a trajectory, which is theoretically more accurate in predicting binding affinity. More computationally intensive molecular dynamics based simulations are linear interaction energy (LIE), thermodynamic integration (TI) or free energy perturbation (FEP). I'd like to use Desmond and have contact with Huafeng, so the particular method we choose depends on its availability in Desmond. Huafeng said that he did binding affinity calculations with Desmond with FEP.
The other issue with a computationally intensive MD simulation is what computational resources do we have. Could I run a job on many nodes of PMC's or Carnegie Mellon's cluster for a week? Alternatively, we could set up Desmond to run in the cloud using Amazon's EC2. We would use the same platform by Cycle Computing that Schrodinger uses. Huafeng said that Desmond should run on the cloud. Cycle Computing takes care of things like data routing, error handling, and various types of automation necessary for running a scientific computing environment on Amazon’s virtual servers.
See the todo list on Aug. 9, 2012 here for summary points on the next steps.
July 31, 2012
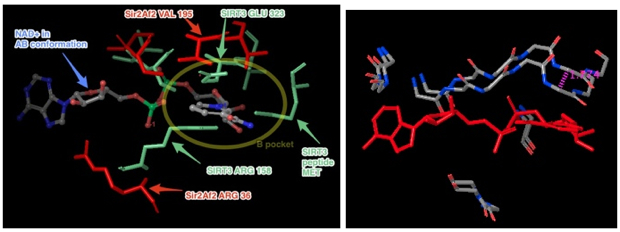
Figures of intermolecular contacts between the Sir2 protein and the nicotinamide of NAD+ in the B and C pockets.
Above figure: There are NO hydrogen bonds between the nicotinamide end of NAD+ in the B pocket and the protein. The 3 shown residues were the only ones within 4 Å of the nicotinamide. The rest of the protein is to the lower left. The B pocket is open to solution above the nicotinamide. The only depicted H-bonds (dashed lines) are not in the B pocket.
Better Figures for Intermolecular Protein-Ligand Interactions: 2D interaction diagrams
The below diagrams are flattened 2D representations of the protein-ligand interactions. In the diagram, residues are represented as colored spheres, labeled with the residue name and residue number. The colors indicate the residue (or species) type:
- red—acidic (Asp, Glu)
- green—hydrophobic (Ala, Val, Ile, Leu, Tyr, Phe, Trp, Met, Cys, Pro)
- purple—basic (Hip, Lys, Arg)
- blue—polar (Ser, Thr, Gln, Asn, His, Hie, Hid)
- light gray—other (Gly, water)
- darker gray—metal atoms
- Solid pink—H-bonds to the protein backbone
- Dotted pink—H-bonds to protein side chains
- Green—pi-pi stacking interactions
- Orange—pi-cation interactions
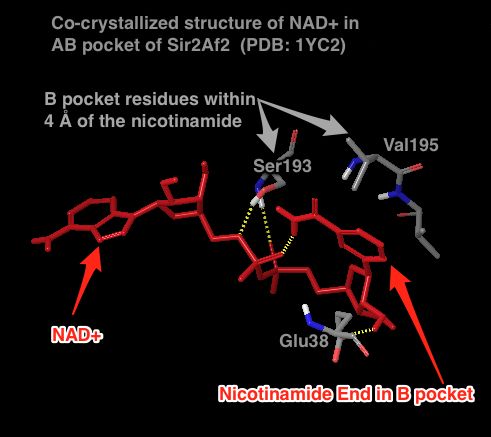
Sir2Af2 (1YC2 chain A) NAD+ in AB pocket
Above figure: Intermolecular protein-ligand diagram of Sir2Af2 (1YC2 chain A) with NAD+ in the AB pocket. Protein and water residues within 3.0 Å of the NAD+ are shown. The lack of a protein "pocket" line around the nicotinamide end and the grey spheres around those atoms indicate that the nicotinamide end is exposed to solvent. The B pocket (show as a yellow circle) is a crevice open to solvent, while the C-pocket (lower yellow circle) is protected by a loop from the solvent (shown in the next diagram). The C-pocket is empty or collapsed in this structure. Also note that there are no H-bonds or other specific intermolecular interactions between the protein and the nicotinamide end of NAD+.
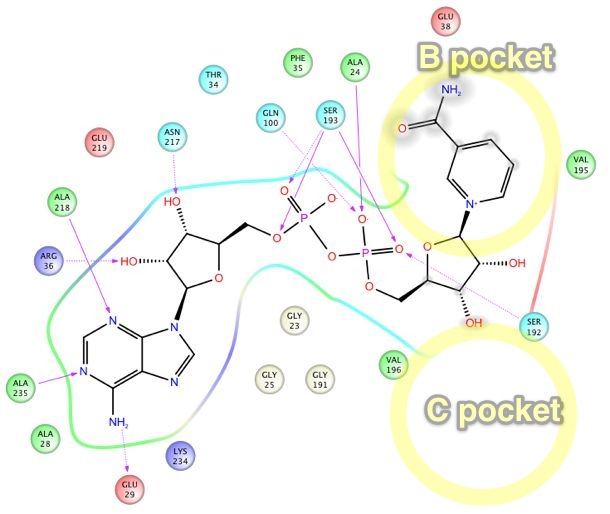
Sir2Af2 (1YC2 chain B) NAD+ in AC pocket

Above figure: Intermolecular protein-ligand diagram of Sir2Af2 (1YC2 chain B) with NAD+ in the AC pocket. Protein and water residues within 3.0 Å of the NAD+ are shown. Unlike in the AB pocket, the NAD+ molecule is completely surrounded by protein residues in the entire A and C pockets. The nicotinamide is not exposed to solvent, unlike in the B pocket. The approximate location of the B pocket is shown due to distortions created by transforming the 3D protein-ligand picture into a simple 2D diagram. There is a backbone H-bond between the protein ILE102 and the nicotinamide end of NAD+.
C pocket for SIRT3 to be added.
July 30, 2012
Sir2 Cross docking:
Some questions have been raised about the results and protocol used for the cross docking studies of Sir2.
The Cross Docking Protocol:
- These are tests in which the protein conformation is from the co-crystallized structure with NAD+ originally in the AC pocket. The NAD+ is removed, and then an attempt is made to re-dock NAD+ into the AB pocket.
- To prevent the NAD+ from re-docking to the AC pocket instead of the AB, an exclusionary volume is placed in the C pocket
- Possibly additional exclusionary volumes surrounding areas neighboring the B pocket are added if the nicotinamide end of NAD+ does not go into the B pocket (as observed from the co-crystallized structure of Sir2 with NAD+ in the AB pocket) with the single exclusion volume in pocket C.
ies were done for
MM-GBSA results for Sir2Af2
Here are the results for in place Glide XP scoring for cross docking of the AB vs. AC conformations of NAD+ from Sir2Af2 (1YC2):
Origin of crystal structures:
- AB conformation: PDB:1YC2:chain D
- AC conformation: PDB:1YC2:chain B
- above AB and AC crystal structures were merged.
- All protein and other molecules were deleted from the AB structure, except for the NAD+ in the AB conformation and a few key waters
- All ligands (NAD+ in AC conformation) and conflicting waters were deleted from the AC structure
- Structure was constrained energy minimized with OPLS2005
- Glide grid was created
- in place scoring with GLIDE XP
- MM-GBSA calculation done from within PRIME.
Vice versa was done for in place docking (scoring) of the NAD+ in the AC pose into the protein structure from the AB conformation.
Results (XP Glide Score):
| Glide XP |
MM-GBSA ∆G bind |
|
| AB NAD+ pose into AC protein |
-11.425 |
-92.553 |
| AC NAD+ pose into AB protein |
-11.220 |
-95.161 |
Discussion
Results likely [[#|agree]] with hypothesis 4 for Sir2.
Hypothesis 4: If NAM is a noncompetitive inhibitor of NAD+, then NAD+ will dock/bind with approximately equal binding affinity to the AB or AC binding pocket. Thus, if NAM occupies pocket C, NAD+ will equally bind to pocket AB.
The two Glide scores for the AB vs. AC binding are very similar, which agrees with the hypothesis. However, the GlideScores are not a reliable indicator for binding affinities or rank ordering the ligands. Thus, we use slightly better measure, the MM-GBSA ∆G_bind, which is calculated as:
dG(1) = E_complex(minimized) - ( E_ligand(minimized) + E_receptor(minimized) )
MM-GBSA is used to estimate relative binding affinity. The absolute values calculated are not necessarily in agreement with experimental binding affinities, thus I am not sure if the units should be kcal/mol. However, the ranking of the ligands based on the calculated binding energies can be expected to agree reasonably well with ranking based on experimental binding affinity, particularly in the case of congeneric series. The two MM-GBSA ∆G_binding of -92.6 and -95.2 are reasonably close to support the original hypothesis 4.
Eric
Comments:
1) The proposal for how to dock to the SIRT3 AB pocket is identical to what I previously saw in an email. If this docking does not work,
the remaining steps do not apply and we will just have those two numbers from MM-GBSA applied to Sir2. As mentioned I will need to see
much more detail in terms of connecting up with the notes from the previous attempts we made, and how things will be done differently in each case this time.
Please connect up as much as possible with the specific details we previously discussed at length.
Also, there are several offline tasks that we discussed that I believe are still pending. I mean the structural superpositions of the various Sir2 and SIRT3 structures with commentary that will allow XG and I to understand which side chains are clashing, which parts of the loop need to be predicted, etc. The latter could be put in the paper irrespective of whether the docking works.
2) Regarding the manuscript, you indicated that you will be placing certain methods descriptions within particular sections of the paper. Why not do some of this now, to whatever extent possible, so we can move along with the manuscript?
3) I agree with the general strategy of motivating MD studies in this paper (and planning out certain aspects of, as you indicated in your wiki task list), but probably not including them in this paper. We will touch base on this again after the pending tasks with docking and induced fit have made more progress.
Raj
Docking Simulations Notes
July 13, 2012
GlideScore vs. MM-GBSA: does the GlideScore contain some implicit solvent calculations?
Short answer: GlideScore does not contain an accurate model for solvation effects to estimate protein-ligand binding affinity, while MM-GBSA estimates solvation effects with a continuum solvent model and a term for solvent accessible surface area.
The GlideScore may contain some solvent effects as describe in the Glide paper (Friesner et al., 2004). The GlideScore does include some explicitly added waters around charged polar groups of the ligand and protein, as well as accounting for possible trapped waters in hydrophobic pockets (which is unfavorable). Glide does this by docking explicit waters into the most favorable poses for the ligand, then scoring these with a "water-score". This description is from the 2004 Glide paper, but it is not well documented in the current Glide manual. I'm not sure if these salvation effects are still in the current 2012 version of GlideScore. Either way, the solvation effects are not similar to how MM-GBSA estimates solvation effects.
MM-GBSA estimates salvation effects differently than the extremely limited solvation estimates in GlideScore (Lyne et al., 2006). MM-GBSA incorporates solvation effects in to the binding energy estimates (ΔEbind):
ΔEbind = ΔEMM + ΔGsolv + ΔGSA Where:
- ΔEMM is the difference in energy between the complex structure and the sum of the energies of the ligand and unliganded protein, using the OPLS force field
- ΔGsolv is the difference in the GBSA solvation energy of the complex and the sum of the solvation energies for the ligand and unliganded protein
- ΔGSA is the difference in the surface area energy for the complex and the sum of the surface area energies for the ligand and uncomplexed protein.
- Corrections for entropic changes are not applied.
Friesner, R.A., Banks, J.L., Murphy, R.B., Halgren, T.A., Klicic, J.J., Mainz, D.T., Repasky, M.P., Knoll, E.H., Shelley, M., Perry, J.K., et al. (2004). Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 47, 1739–1749.
Lyne, P.D., Lamb, M.L., and Saeh, J.C. (2006). Accurate Prediction of the Relative Potencies of Members of a Series of Kinase Inhibitors Using Molecular Docking and MM-GBSA Scoring. J. Med. Chem. 49, 4805–4808.
July 13, 2012
Plan for continued simulations
- MM-GBSA calculations for static docking poses of Sir2 from the previous results. Recall that the previous calculated docking energies agreed with the what was expected from experiment. However, publishing the docking energies would not be defendable in a paper, while MM-GBSA energies have at least one leg to stand on. These calculations will not take long.
- SIRT3: getting docking to work. How? A number of tweaks or alternative steps could work. I just need to find the correct local energy minimum structure for the AB pocket docking of NAD+. Once we have that, subsequent simulations can start from this structure. A few possibilities:
- adjust the soft core potential to allow closer contacts so docking can occur in the AB pocket
- Do a more comprehensive testing of induced fit. Previously I tried a handful of different setups. This time I could try more. Recall how Brad got DOCK to successfully work for some test cases - he systematically adjusted the parameters to minimize the RMSD to the crystal structure. A similar strategy could work here.
- Do a constrained minimization, where the NAD+ is kept frozen in the AB pocket, and allow the protein to adjust around it. Subsequently use PRIME to optimize the side chains for this new conformation. Compare the backbone to the starting structure and to other Sirtuins co-crystallized in the AB conformation.
- After successfully docking NAD+ in the AB pocket of SIRT3, a number of the hypothesis can be tested, as previously described in detail in the Sirtuin Kinetics paper draft I wrote (DropBox:PMC-AT Research / Eric / Sirtuin Kinetics Paper draft 5-16-2012.doc) Many of these involve the difference in the calculated binding energy of NAD+ between the AB and AC pockets, as well as effects that the nicotinamide inhibitor has on those differences. In particular, how will these new simulations connect with the previously planned simulations described in the document?
- Hypothesis 1a for SIRT3 can now be done. This is a simulation to test if NAD+ preferentially docks to AC pocket vs. AB pocket. Now, with the docking results from SIRT3 with NAD+ in the AB pocket and the AC pocket, and subsequent MM-GBSA calculations on these docked complexes, the MM-GBSA binding affinity estimate between the AB and AC poses will be compared. The hypothesis here is that the binding affinity is higher for AC than for AB.
- Hypothesis 1b is similar to 1a, but for Sir2 instead of SIRT3.
- Hypothesis 2 and 3: these test if there are any structural changes induced by the nicotinamide inhibitor upon binding to the C pocket, which would affect the binding affinity of NAD+ to the AB pocket. Docking protocols alone will not answer this question. A full MD method would be best to model protein structure changes upon binding of an inhibitor. An intermediate modeling approach proposed here involves using docking and side chain / loop prediction modeling (PRIME + Glide = Induced Fit). 1st the inhibitor is docked into the C pocket. Then, side chain and loop minimization with Prime relaxes the inhibitor-protein complex. Finally, NAD+ will be re-docked to the AB pocket to find how the inhibitor-protein complex affects binding of the NAD+.
- Hypothesis 6 and 7 are about where and how various inhibitors dock. Some work has been previously done on these hypothesis. More work here will be cleaning up and summarizing results, as well as a few extra simulations.
- Paper: you asked for a break down of where in the paper the computational sections will go. Results and methods will describe the simulation details, while a section of the discussion will describe the benefits and limitations of the docking simulations with the MM-GBSA. In the area describing further research, a few remarks of future work to address those limitations will discuss more advanced molecular dynamics based techniques.
- Docking of a small collection of inhibitors that Xiangying has tested or plans to test. I cannot dock a large library of inhibitors, but I can dock a smaller library of dozens to thousands. There is a list of inhibitors that would be interesting to dock and calculate binding energies with MM-GBSA.
- Plan and execute a molecular dynamics simulation to calculate binding energy. No details here yet. Only pursue after complete above.
July 3, 2012
Discussion: How to correlate protein-ligand docking scores or other more advanced simulations techniques to binding affinity?
A presentation is available here:
[[#|DropBox]]:PMC-AT Research/Eric/binding.affinity.predictions.ppt
The notes (not ordered) for the power point presentation are here:
[[#|DropBox]]:PMC-AT Research/Eric/binding.affinity.predictions.doc
July 3, 2012 meeting notes
Present on conference call: Raj, Xiangying, Eric
Title: Binding Affinity Predictions
- The MM-GBSA technique to improve a protein-ligand docking score is worth trying as a next step with the previously docking studies on sirtuin proteins
- MM-GBSA is available in the Schrodinger software suite.
- MM-GBSA is a continuum solvent model (Generalized Bourne solvent model combined with a surface area technique). This technique is an improvement of over standard Glide, because Glide does not properly account for solvation effects.
- Raj: Does Prime use GBSA or SGB by default?
- the special protocol of MM-GBSA to predict binding affinities and improve the GlideScore uses GBSA, but we do not know if the GBSA protocol is also used in standard PRIME simulations for side chain and loop predictions
- Reorganization energy: how is this accounted for?
- in Glide
- in Induced Fit
- the energy change from moving loops or side chains in the protein to optimize the protein-ligand conformation may be an important contribution to the overall binding affinity. It is not clear how Glide or Induced Fit account for this. Every docking result has both a GlideScore and an eModel score. How is this eModel score used and does it properly represent reorganization energy? Is the eModel score also used with the GlideScore to rank order results?
- Find a reference for how scoring functions are designed
- specifically, Eric commented that scoring functions in protein-ligand docking programs such as Glide and DOCK are optimized to minimize the RMSD of the docked ligand compared to a crystal structure. The experimentally determined structures are from a large training set of co-crystallized protein-ligand x-ray structures. Although there is probably some correlation between optimizing to match crystal structure ligand poses and binding affinity, the scoring functions are NOT optimized to give the best binding affinity predictions.
- Many of the presented techniques for predicting binding affinity do the following:
- Step (1) do a standard docking simulation with Glide or other program
- Step (2) post-docking scoring with some better / newer scoring scheme than from (1), but uses the same pose predicted in (1).
- Raj pointed out that this may be a limitation. What if the pose from (1) is wrong? Well, the improvement from (2) is predicated on the standard docking pose from (1) being close to correct. Yes, this could be a limitation, as it realies completely on pose sampling from (1) rather than the improved scoring scheme from (2). Still, this post-docking re-scoring is a common possibly because using the scoring theme from (2) may not be the best way to find the best poses.
- How to address above limitation when doing MM-GBSA to get better binding affinity predictions?
- For example, if we do Glide or Induced Fit docking, then run a MM-GBSA calculation on those found docked ligand poses: will the new MM-GBSA scores change the rank ordering of poses for a given ligand as compared to the rank ordering from the GlideScore alone?
- How can we explain this? Use the argument that the GlideScore alone does not properly account for solvation effects. While the MM-GBSA is a very simple continuum solvent model, it does make a basic attempt at including solvation affects, which have been shown to be very important for many protein-ligand complexes.
- What if the GlideScore ranked poses omit the correct pose? Then subsequent re-scoring with MM-GBSA will NOT necessarily improve the binding affinity prediction. Because of this, each ligand should be liberally sampled with the initial Glide docking, and every output pose from the top of the rank ordered list to the bottom should be re-scored with MM-GBSA.
- Raj: Why are the freely available DOCK and AUTODOCK not on the table 2 from
- H. Ashtawy and N. Mahapatra, “A Comparative Assessment of Ranking Accuracies of Conventional and Machine-Learning-Based Scoring Functions for Protein-Ligand Binding Affinity Prediction,” Computational Biology and Bioinformatics, IEEE/ACM Transactions on, vol. PP, no. 99, p. 1, 2012.
- This table compared Glide and GOLD and other techniques against the PDBbind benchmark
- Can we use the RF-score from the Mahapatra 2012 out of the box for our sirtuin systems?
- yes, probably. That is, we do not re-optimize this machine learning scoring technique specifically for our sirtuin system. We use it with the same parameters as found in the paper when trained against the PDBbind data set.
- Difference between comparing binding affinities in different modes vs. a cogeneric series of molecules?
- many of the presented studies optimized a scoring function for a series of cogeneric molecules docking in the same pocket.
- an important question that we are studying is different: what is the difference in binding affinity of the same molecule to two different binding poses (AB vs. AC pockets).
- Eric: techniques such as MM-GBSA or others that improve scores may still be helpful, but we must be aware of the subtle difference.
- TO DO:
- Get academic license of Desmod for molecular dynamics simulations
- email Huafeng Xu to facilitate further discussions with him about MD simulations
- figure out what to do next about the SIRT3 docking studies which did not work.
- list docking runs that will do with MM-GBSA